Your basket is currently empty!
B Cell Linear Epitope Mapping
B cell epitope mapping
a straightforward way for characterizing B cell epitopes in any project
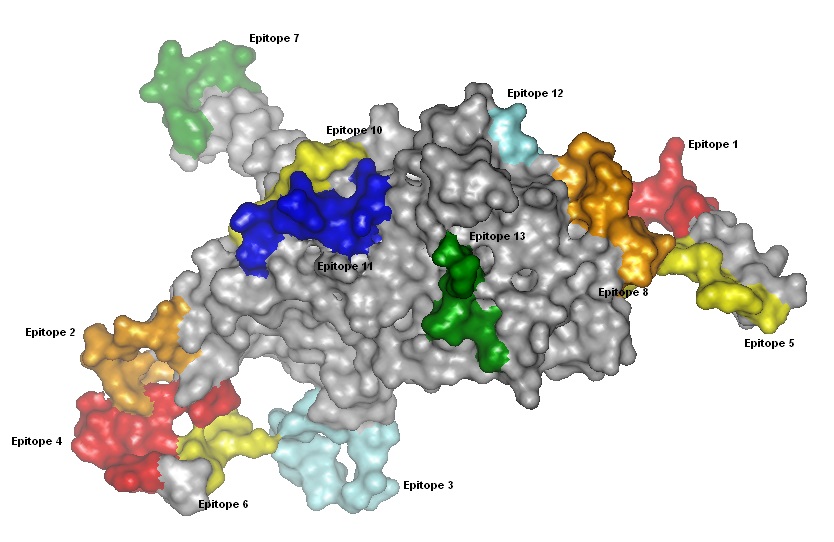
Figure 1. B cell epitopes of Human papilloma virus 16 major late capsid protein L1
Do you need to analyze antibodies in serum samples, or identify the specific binding region for a monoclonal antibody?
Are you trying to select the best monoclonal antibody from a panel by characterizing strength of binding, or target specificity?
Setting up assays in-house to test a large number of samples or screen many epitopes can be fraught with problems, such as the high cost of specialist equipment, inexperienced operators and a lack of resources to set up and maintain the assays.
Outsource your B cell linear epitope mapping to the experts
ProImmune’s service provides mapping of B cell linear epitopes in any protein or polypeptide sequence using antibody or antisera samples submitted by the client. The specific linear peptide segments from any larger protein that bind to antibodies in a given sample can be identified using immunoassay technology. The assay uses high-quality peptides in 96-well plate format.
Our services include cost-effective solutions for a wide range of project requirements, for example, from a handful of peptides screened against a similar number of antibody samples, to hundreds or even thousands of peptides screened against hundreds of antisera.
Mapping linear epitopes can be performed quickly with well-defined, easily controllable, high-throughput technologies. Although not all conformational epitopes are resolved, it can be a quick way to achieve relative ranking of antigenic regions within proteins and between different proteins in a group.
If the antibody-binding region of the protein is unknown, an overlapping peptide library can be generated using a specific peptide length and offset, for example, 15mers offset by 5 amino acids, i.e. overlapping by 10. Using our epitope mapping service, the specific linear protein segments from any larger protein or peptide, that bind to antibodies in a given sample such as sera or plasma, can be identified. In this way, hot-spot areas of the protein can be discovered (for example, see figure).
|
|
|
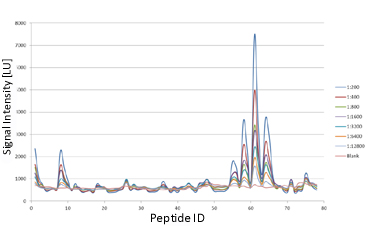
| Figure 2. Use of an overlapping peptide library to map epitopes from HBV surface antigen. Graph shows signals obtained with decreasing dilutions of patient serum tested at ProImmune |
Alternatively, if these areas of the protein sequence are already defined, the optimal epitope sequence can be elucidated using with a peptide library with an offset of 1, or with a truncated peptide library, or with a series of peptides with single amino acid substitutions; see Peptide Library Design.
Data generated from the assay will depend upon the quality of the antibody being tested. It should be noted that sera and plasma samples sometimes contain multiple antibodies, which may give weak signals and/or bind to several different peptides. The data generated from such samples can still provide good information about which regions of a protein sequence are most antigenic, and highlight areas on which to carry out further investigation. Discontinuous epitopes that depend on the correct 3D conformation of the protein, may not be detected by linear epitope mapping, or may only give a weak signal.
Our customer service team of PhD immunologists and biochemists can add real value to your research projects. Contact us to discuss your project and we will design an assay and array format tailored to achieve your objectives.
Key Publication:
Arnaboldi PM, et al. (2013). “Outer Surface Protein C Peptide Derived from Borrelia burgdorferi Sensu Stricto as a Target for Serodiagnosis of Early Lyme Disease”. Clin Vaccine Immunol. 20(4):474-81. [PubMedID: 23365204]
(read the full case study here)
Questions that can be addressed using B cell linear epitope mapping
Definition and ranking of individual linear epitopes in an antigen
Although protein antigens may contain both linear and discontinuous epitopes, detailed knowledge of the linear epitopes alone may be a useful means of comparing the antigenicity of candidate proteins.
Identification of peptides for immunization
If a strong candidate linear epitope can be found in a protein antigen, this short peptide sequence can be used to produce polyclonal or monoclonal antibodies against the target protein, with the advantage that the binding sequence in the protein will already be known.
Discovery of cross-reactive determinants between related proteins
Determining a linear epitope that occurs on several related proteins gives useful information about the potential cross-reactivity of immune responses between these proteins.
Definition of strongly antigenic regions in proteins
Clusters of linear epitopes may indicate regions of elevated antigenicity and activity in a protein and may be useful in identifying protein sub-units relevant to the research application. For example, a sub-unit of a protein may confer most of the antigenicity of the whole protein and be easier to manufacture than the whole protein. In addition, to reduce the risk of unwanted side effects in vaccination it may be of benefit to use a sub-unit of a protein that has fewer epitopes that are cross-reactive with self-proteins.
Comparison of different serovars or subtypes in terms of their relevant antigenicity
Comparative results from linear mapping using peptides with sequence variation can shed light on the antigenicity of proteins from different strains of an organism. This application may also show whether vaccination produces an immune response that is relevant across a number of serovars or subtypes.
Combining linear epitope mapping with epitope prediction
Comprehensive B cell epitope prediction and linear epitope mapping can be carried out when both the 3D structure and the protein sequence are known. However, the 3D structure is not available for many proteins, especially transmembrane proteins, which are particularly difficult to crystallize. In vaccine development, transmembrane proteins are of special interest when they have a domain on the surface of the pathogen that could be bound by antibodies. Linear epitope mapping alone, or combined with epitope prediction, can give an insight into the antigenicity of transmembrane proteins.
Characterizing the contribution of individual amino acids to the binding interaction
An alanine scanning peptide library can be used to discover amino acids in the sequence that are essential to binding. Alanine is sytematically substituted into each position in the epitope to identify the amino acids that are essential for the binding interaction with the antibody or protein.