Your basket is currently empty!
Pentamers-are-used
Case Study:
Pro5® MHC Pentamers are used to identify a new target for EBV treatment
Fox, CP., et al. (2010). A novel latent membrane 2 transcript expressed in Epstein-Barr virus-positive NK- and T-cell lymphoproliferative disease encodes a target for cellular immunotherapy. Blood. 116(19): 3695-704. [PubMed ID: 20671118]
In an issue of Blood, Christopher Fox and his collaborators explain a mystery from the EBV field.
Natural Killer/T cell lymphomas are frequently associated with EBV and respond poorly to current therapeutic regimes. The EBV genome encodes some 80 proteins, but very few of these are expressed in tumours. Among these few is the membrane protein LMP2. LMP2 is targeted by CD8+ T cells (CTLs) and shows some promise as an avenue of attack for anti-tumour therapy.
Fox et al characterized four EBV tumour cell lines, and key to their analysis was the range of LMP2-specific MHC Class I Pro5® Pentamers available from ProImmune, which they used in flow cytometry. They found that LMP2A was barely detectable as a protein (using an antibody specific for the unique N-terminal of the ‘A’ variant), and using primers from the 5’ end of each transcript found vanishingly small levels of each. Given that anti-LMP2 CTL responses have been previously characterized, this result was baffling.
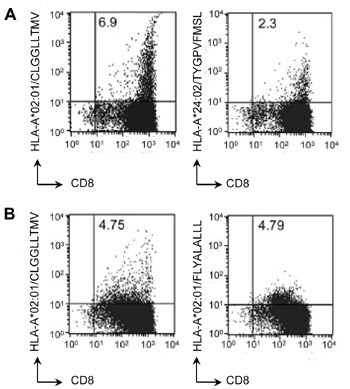
Importantly, antigen presentation by the cell lines was intact. The team made two different polyclonal anti-LMP2 CTL preparations, by stimulating EBV patient PBMC with LMP2-transduced antigen presenting cells. The polyclonal CTLs were active against tumour cell lines, as measured by 51-Cr release assay. Their CTL mixture contained substantial numbers of LMP2-specific cells, identified by Pro5® Pentamer staining (Figure 1).
|
|
Figure 1: Dual-color flow cytometry of CTLs derived from two EBV-associated lymphoma patients (A and B) prepared by ex vivo stimulation with LMP2/LMP1 transfected antigen-presenting cells. CTLs were stained with anti-CD8 antibody and with HLA-peptide pentamers: HLA-A*02:01/CLGGLLTMV, HLA-A*02:01/FLYALALLL, HLA-A*24:02/TYGPVFMSL as indicated. Numbers in the top right quadrant of each plot indicate the proportion of viable peptide/pentamer-specific CD8+ cells as a percentage of viable cells. |
Since activity against other EBV epitopes was not excluded by this analysis, A*24:02 and A*02:01 epitope-specific clonal CTL lines were also tested, and gave comparable results. Widening the HLA scope of their investigation, Fox et al transduced one of their tumour cell lines with recombinant HLA-A*11:01, and observed IFNg production from a HLA-A*11:01-restricted LMP2 epitope-specific CTL clone.
So, with anti-LMP2 CTL responses in the context of three different HLA types, in the apparent absence of LMP2 expression – could there be more transcripts from the LMP2 locus? qRT-PCR of 3’ exons of the LMP2 locus showed robust expression of a transcript encoding (at least) exons 2-6. Via 5’-RACE, the new transcripts were found to initiate in the terminal repeat (TR) region of the LMP2 gene, and all four EBV tumour cell lines tested expressed transcripts initiating in the TR.
This result was validated using mRNA extracted from patient biopsies. Of seven EBV-associated lymphomas examined, all expressed the newly-identified transcript. The implications of this finding are far-reaching, as the so-called TR-LMP2 protein, being the only LMP2 variant expressed during NK and T-cell lymphomagenesis, could become a target for future therapies. The ability to map individual epitope responses from CTL, using ProImmune Pentamers, was critical to the success of this work.
ProImmune Pro5® Pentamers Used:
| F683-2A | A*24:02/TYGPVFMSL | EBV LMP2 (419–427) |
| F666-2A | A*02:01/FLYALALLL | EBV LMP2 (356-364) |
| F508-2A | A*11:01/SSCSSCPLSK | EBV LMP2 (340-349) |
| F042-2A | A*02:01/CLGGLLTMV | EBV LMP2 (426-434) |
This work was carried out in the Tumour Immunology and Gene Therapy labs in the University of Birmingham School of Cancer Sciences, in collaboration with the Bollard laboratory at Baylor College of Medicine, Houston, Texas (shown below).

Data reproduced with permission of AMERICAN SOCIETY OF HEMATOLOGY (ASH) via Copyright Clearance Center.